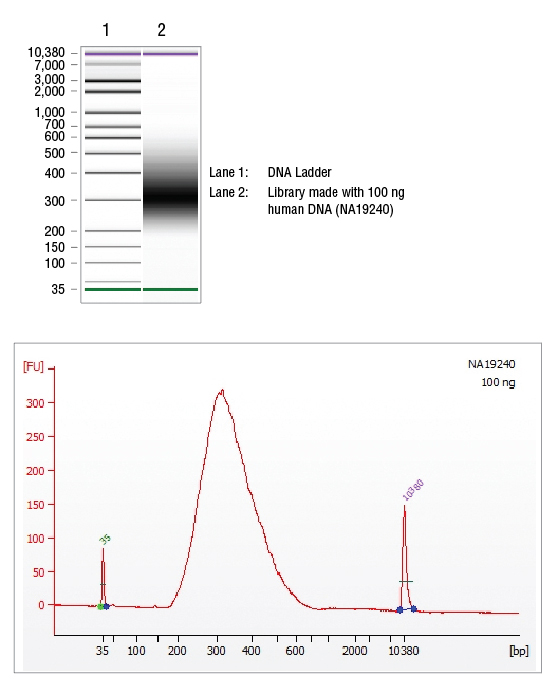
Protocol for NEBNext Direct® Cancer HotSpot Panel (NEB #E7000)
Critical Guidelines
• Note that the captured molecules on streptavidin beads are unamplified, so any beads lost to pipette tips will result in fewer unique targets in the final library and reduced overall yields. Take care to minimize bead loss during washes.
• Perform all washes by pipetting up and down at least 10 times. Insufficient washing can lead to carryover of previous reagents, resulting in adaptor dimer formation and reduced yields. Foaming of the wash buffers may occur due to the presence of detergents.
• A thermocycler programmable to 100 μl is required for this protocol. A thermocycler programmable to only 50 μl is not compatible with the incubations. The hybridization, bead binding, and wash volumes will exceed 100 μl, but a thermocycler programmable to 100 μl is sufficient for these steps.
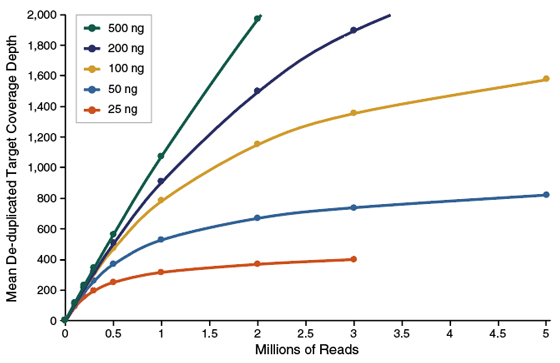
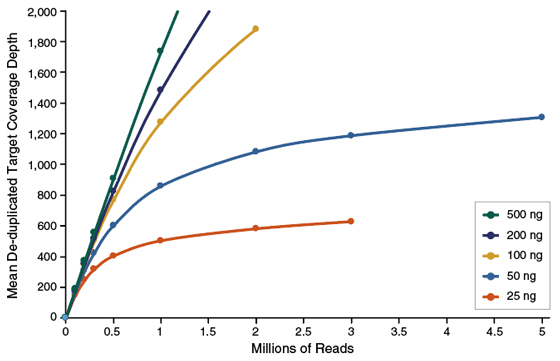
• NEBNext Direct Cancer HotSpot panel is compatible with either PE75 or PE150 Illumina sequencing. Please refer to Figure 1.2 (at the end of the protocol examples) for examples of typical coverage depths across DNA input amounts with PE75 (Figure 1.2A) and PE150 (Figure 1.2B) sequencing reads.
• Note that the Post-ligation Washes (Sections 1.8.7 and 1.10.7) contain different steps than the Post-reaction Washes (Sections 1.6.6, 1.7.6, 1.9.7 and 1.11.7).
• This protocol is incompatible with the Illumina Experiment Manager. For guidelines on how to run samples on an Illumina MiSeq, see Chapter 4.
• Reaction temperatures are carefully optimized for best target conversion. Strictly adhere to reaction temperatures and times throughout the protocol.
Data Processing
• For information on bioinformatic utilization of unique molecular identifiers (UMIs) please refer to Using Unique Molecular IDs with NEBNext Direct – Data Usage Guideline Page within the "Usage Guidelines & Tips" panel in the "Other Tools & Resources" tab at: www.neb.com/E7000.
• We have developed and optimized a pipeline for processing of data directly from FASTQ files using open-source bioinformatics tools. Details on this pipeline can be found at: https://github.com/DirectedGenomics/DemoPipeline.
• The i5 UMI index is recorded as a series of 12 Ns for the sequencing run. Please confirm that the BCL to FASTQ converter to be used tolerates Ns in the reported i5 index sequence prior to analysis.
Helpful Tips
• If you are preparing more than one sample, we recommend making master mixes using ~10% more volume than required per reaction.
• A multichannel pipette (200 μl) is recommended for washing and mixing steps if preparing multiple samples.
• To save time, buffers for all master mixes in steps 1.6–1.11 (3´ Blunting through Adaptor Cleaving) can be pre-aliquoted and kept at 4°C until needed. Enzyme can be added to the buffers during the previous incubation and kept on ice until needed.
• It is ideal to use a thermocycler on a bench with accessible workspace for this protocol.
Before You Begin
• Bring all wash solutions and beads (Box 2) to room temperature before use.
• Bead Wash 1 (BW1) may contain a precipitate due to shipping at 4°C. This is normal and should go back into solution once the buffer reaches room temperature (at least 4 hours followed by gently inverting the bottle ~10 times). Do not use until all precipitate has dissolved. If necessary, incubate the bottle in a 25°C or 37°C incubator or water bath to dissolve the precipitate.
Protocol for NEBNext Direct Cancer HotSpot Panel
Symbols| This caution sign signifies a step in the protocol that has multiple paths leading to the same end point but is dependent on a user variable, like the amount of input DNA. | |
| Stopping points in the protocol. | |
 |
Colored bullets indicate the cap color or label stripe of the reagent to be added to a reaction. |
1.1. Quantify DNA
1.1.1. Quantify input DNA using a Qubit™ dsDNA HS Assay Kit or a Quant-iT™ PicoGreen™ dsDNA Assay Kit. Do not use a UV-based assay for quantification.
1.2. DNA Nicking
![]() If you are starting with cell-free DNA, skip DNA Nicking and go directly to Bait Hybridization in Section 1.3.
If you are starting with cell-free DNA, skip DNA Nicking and go directly to Bait Hybridization in Section 1.3.
![]() If you prefer to fragment with Covaris®, skip DNA Nicking and shear input DNA in 52 μl (total volume) of 1X TE. Follow Covaris recommendations for instrument and microtube setup using the 200 bp target size protocol for shearing in 50 μl. Transfer 50 μl of the sheared DNA to a PCR well, then proceed directly with bait hybridization in Section 1.3.
If you prefer to fragment with Covaris®, skip DNA Nicking and shear input DNA in 52 μl (total volume) of 1X TE. Follow Covaris recommendations for instrument and microtube setup using the 200 bp target size protocol for shearing in 50 μl. Transfer 50 μl of the sheared DNA to a PCR well, then proceed directly with bait hybridization in Section 1.3.
1.2.1. Take out the stop solution to thaw.
1.2.2. Set up the following reaction on ice. First, mix the DNA, buffer and water in a PCR tube or well. Add the enzyme last.
| REAGENT | PER REACTION |
|---|---|
| DNA (10-1,000 ng)* | 1-38 μl |
| 4 μl | |
| 3 μl | |
| Ice-cold, nuclease-free H2O | variable |
| Total volume | 45 μl |
* For detection of somatic variants, we strongly recommend starting with a minimum of 100 ng DNA input.
1.2.3. Mix the reaction thoroughly by pipetting up and down 10 times. Incubate at 18°C for 15 minutes. Reactions may be incubated capped or uncapped.
1.2.4. Stop the reaction by moving the samples to ice and adding 5 μl of ![]() (brown) stop solution. Mix thoroughly by pipetting up and down 10 times.
(brown) stop solution. Mix thoroughly by pipetting up and down 10 times.
1.2.5. Proceed directly to Bait Hybridization in Section 1.3.
Note that at this stage, the DNA is nicked, not fragmented. Therefore, these reactions cannot be analyzed for size as dsDNA on non-denaturing gels or on Agilent’s TapeStation, Bioanalyzer or similar instruments.
1.3. Bait Hybridization
1.3.1. Make a hybridization master mix by adding the following components for the appropriate number of reactions. Vortex the hybridization buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
| 47 μl | 51.7 μl | |
| 20 μl | 22 μl | |
| 5 μl | 5.5 μl | |
| Total | 72 μl | 79.2 μl |
1.3.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.3.3. To each sample of nicked or fragmented DNA from Section 1.2, add 72 μl of hybridization master mix for a final volume of 122 μl. Mix by pipetting up and down 10 times. Seal the PCR plate or cap tubes securely to avoid evaporation.
1.3.4. Run the following program with the heated lid set to 105°C and place the samples in the thermocycler after the block temperature reaches 95°C:
10 mins @ 95°C
90 mins @ 60°C
Hold @ 60°C
1.3.5. While the samples are incubating, prepare  (blue) Streptavidin beads (see Streptavidin Bead Preparation in Section 1.4).
(blue) Streptavidin beads (see Streptavidin Bead Preparation in Section 1.4).
1.3.6. After the incubation at 60°C and when Section 1.4 (Streptavidin Bead Preparation) is complete, unseal the tubes/wells, leave the samples on the thermocycler at 60 C with the lid open and proceed to Bead Binding in Section 1.5.
1.4. Streptavidin Bead Preparation
1.4.1. Warm Streptavidin beads to room temperature (~15 minutes).
1.4.2. Vortex the Streptavidin beads to resuspend.
1.4.3. For each reaction, 75 μl of beads are required (82.5 μl with 10% overage). In a 2 ml Eppendorf tube, add the appropriate volume of beads for the number of reactions performed.
Note: Use multiple 2 ml tubes if performing more than 12 reactions. It is not recommended to exceed 1 ml of beads per 2 ml Eppendorf tube.
1.4.4. Place the tube(s) on a magnet and wait for the solution to clear (~1 minute). Remove the supernatant, and then remove the tube(s) from the magnet.
1.4.5. Add 150 μl of  (blue) Hybridization Wash (HW) per reaction (165 μl with 10% overage) to the beads and resuspend by vortexing or pipetting.
(blue) Hybridization Wash (HW) per reaction (165 μl with 10% overage) to the beads and resuspend by vortexing or pipetting.
1.4.6. Place the tube(s) on a magnet and wait for the solution to clear (~1 minute). Remove the supernatant, and then remove the tube(s) from the magnet.
1.4.7. Repeat Steps 1.4.5–1.4.6 twice for a total of 3 washes.
1.4.8. Resuspend the beads in 30 μl of  (blue) Bead Prep Buffer per reaction (33 μl with 10% overage).
(blue) Bead Prep Buffer per reaction (33 μl with 10% overage).
1.4.9. Keep the beads at room temperature until bait hybridization (Section 1.3) is completed.
Note: For Sections 1.5–1.11, the thermocycler lid should be open and unheated. The PCR tubes can remain uncapped for ease of mixing. However, if it is preferred, tubes can be capped during incubations.
1.5. Bead Binding
1.5.1. Immediately before use, vortex the washed Streptavidin Beads (from Step 1.4.9) in Bead Prep Buffer to resuspend.
1.5.2. Add 30 μl of resuspended beads to each reaction (from Step 1.3.6) while the samples are on the thermocycler at 60°C, and then mix gently by pipetting up and down 10 times.
Note: If you have a large number of samples, the Streptavidin Beads may start to settle as you distribute the beads to each reaction. To prevent this, pipette up and down or vortex the beads frequently to keep them in suspension while pipetting into individual reactions.
1.5.3. Change the thermocycler temperature to 48°C and incubate the reactions for 10 minutes.
1.5.4. Remove the samples from the thermocycler and place on a magnet. Wait for the solution to clear (~15 seconds), remove the supernatant, and then remove the samples from the magnet.
1.5.5. Add 150 μl of HW to each sample. Mix by pipetting up and down 10 times. Place the samples on a thermocycler set at 62°C (lid open) and incubate for 5 minutes.
1.5.6. Remove the samples from the thermocycler and place on a magnet. Wait for the solution to clear (~15 seconds), remove the supernatant, and then remove the samples from the magnet.
1.5.7. Repeat Steps 1.5.5–1.5.6 for a total of 2 washes at 62°C.
1.5.8. Add 150 μl of ![]() (light pink) Bead Wash Buffer 2 (BW2) to each sample. Mix by pipetting up and down 10 times.
(light pink) Bead Wash Buffer 2 (BW2) to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 3´ Blunting of DNA in Section 1.6.
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 3´ Blunting of DNA in Section 1.6.
1.6. 3´ Blunting of DNA
1.6.1. While the beads are suspended in BW2 buffer, make a 3´ Blunting master mix by adding the following components in a sterile nuclease-free tube for the appropriate number of reactions. Vortex the 3´ Blunting Buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
| 97 μl | 106.7 μl | |
| 3 μl | 3.3 μl | |
| Total | 100 μl | 110 μl |
1.6.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.6.3. Place the DNA-bound beads on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.6.4. Add 100 μl of 3´ Blunting master mix (from Step 1.6.1) to each sample. Gently mix by pipetting up and down 10 times. Incubate the samples at 37°C for 10 minutes on a thermocycler with the thermocycler lid open.
Note: For optimal performance, it is important that the incubation time for this step is closely followed.
1.6.5. Proceed immediately with the Post-reaction Wash (Section 1.6.6).
1.6.6. Post-reaction Wash
1.6.6.1. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.6.6.2. Add 150 μl of ![]() (dark purple) Bead Wash Buffer 1 (BW1) to each sample. Mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
(dark purple) Bead Wash Buffer 1 (BW1) to each sample. Mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.6.6.3. Add 150 μl of BW2 to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA – bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to dA-Tailing in Section 1.7.
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA – bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to dA-Tailing in Section 1.7.
1.7. dA-Tailing
1.7.1. While the beads are suspended in BW2 buffer, make a dA-Tailing master mix by adding the following components in a sterile nuclease-free tube for the appropriate number of reactions. Vortex the dA-Tailing Buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
| 97 μl | 106.7 μl | |
| 3 μl | 3.3 μl | |
| Total | 100 μl | 110 μl |
1.7.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.7.3. Place the DNA-bound beads on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.7.4. Add 100 μl of dA-Tailing master mix (from Step 1.7.1) to each sample. Gently mix by pipetting up and down 10 times. Incubate the reactions at 37°C for 10 minutes on a thermocycler with the thermocycler lid open.
1.7.5. Proceed immediately with the Post-reaction Wash (Section 1.7.6).
1.7.6. Post-reaction Wash
1.7.6.1. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.7.6.2. Add 150 μl of BW1 to each reaction and then mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~ 15 seconds). Remove the supernatant, and then remove the reactions from the magnet.
1.7.6.3. Add 150 μl of BW2 to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 3´ Adaptor Ligation in Section 1.8.
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 3´ Adaptor Ligation in Section 1.8.
1.8. 3´ Adaptor Ligation
1.8.1. While the beads are suspended in BW2 buffer, make a 3´ Adaptor Ligation master mix by adding the following components in a sterile nuclease-free tube for the appropriate number of reactions. Vortex the Adaptor Ligation Buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
| 80 μl | 88 μl | |
| 10 μl | 11 μl | |
| 10 μl | 11 μl | |
| Total | 100 μl | 110 μl |
1.8.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.8.3. Place the DNA-bound beads on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.8.4. Add 100 μl of 3´ Adaptor Ligation master mix (from Step 1.8.1) to each sample. Gently mix by pipetting up and down 10 times.
1.8.5. Incubate the samples at 20°C for 15 minutes on a thermocycler with the thermocycler lid open.
1.8.6. Proceed immediately with the Post-ligation Wash (Section 1.8.7).
1.8.7. Post-ligation Wash
Note: The following wash steps are different than the Post-reaction Washes.
1.8.7.1. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.8.7.2. Add 150 μl of BW1 to each sample. Mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.8.7.3. Repeat Step 1.8.7.2 for a total of 2 washes in BW1.
1.8.7.4. Add 150 μl of BW2 to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 5´ Blunting of DNA in Section 1.9.
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 5´ Blunting of DNA in Section 1.9.
1.9. 5´ Blunting of DNA
1.9.1. While the beads are suspended in BW2 buffer, make a 5´ Blunting master mix by adding the following components in a sterile nuclease-free tube for the appropriate number of reactions. Vortex the 5´ Blunting Buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
 (orange) 5´ Blunting Buffer (orange) 5´ Blunting Buffer |
97 μl | 106.7 μl |
 (orange) 5´ Blunting Enzyme Mix (orange) 5´ Blunting Enzyme Mix |
3 μl | 3.3 μl |
| Total | 100 μl | 110 μl |
1.9.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.9.3. Place the DNA-bound beads on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.9.4. Add 100 μl of 5´ Blunting master mix (from Step 1.9.1) to each sample. Gently mix by pipetting up and down 10 times.
1.9.5. Incubate the samples at 20°C for 10 minutes on a thermocycler with the thermocycler lid open.
1.9.6. Proceed immediately with the Post-reaction Wash (Section 1.9.7).
1.9.7. Post-reaction Wash
1.9.7.1. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.9.7.2. Add 150 μl of BW1 to each sample. Mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.9.7.3. Add 150 μl of BW2 to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds).
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds).
Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to 5´ Adaptor Ligation in Section 1.10.
1.10. 5´ Adaptor Ligation
1.10.1. While the beads are suspended in BW2 buffer, make a 5´ Adaptor Ligation master mix by adding the following components in a sterile nuclease-free tube for the appropriate number of reactions. Vortex the Adaptor Ligation Buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
| 80 μl | 88 μl | |
| 10 μl | 11 μl | |
| 10 μl | 11 μl | |
| Total | 100 μl | 110 μl |
1.10.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.10.3. Place the DNA-bound beads on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.10.4. Add 100 μl of 5´ Adaptor Ligation master mix (from Step 1.10.1) to each sample. Gently mix by pipetting up and down 10 times.
1.10.5. Incubate the samples at 20°C for 20 minutes on a thermocycler with the thermocycler lid open.
1.9.6. Proceed immediately with the Post-ligation Wash (Section 1.10.7).
1.10.7. Post-ligation Wash
Note: The following wash steps are different than the Post-reaction washes.
1.10.7.1. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.10.7.2. Add 150 μl of BW1 to each sample. Mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.10.7.3. Repeat Step 1.10.7.2 for a total of 2 washes in BW1.
1.10.7.4. Add 150 μl of BW2 to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to Adaptor Cleaving in Section 1.11.
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to Adaptor Cleaving in Section 1.11.
1.11. Adaptor Cleaving
1.11.1. While the beads are suspended in BW2 buffer, make a Cleaving master mix by adding the following components in a sterile nuclease-free tube for the appropriate number of for the appropriate number of reactions. Vortex the Cleaving Buffer to mix well prior to pipetting.
| REAGENT | PER REACTION | WITH 10% OVERAGE |
|---|---|---|
 (lilac) Cleaving Buffer (lilac) Cleaving Buffer |
95 μl | 104.5 μl |
 (lilac) Cleaving Enzyme Mix (lilac) Cleaving Enzyme Mix |
5 μl | 5.5 μl |
| Total | 100 μl | 110 μl |
1.11.2. Mix the master mix well by vortexing for 3–5 seconds and centrifuge briefly.
1.11.3. Place the DNA-bound beads on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.11.4. Add 100 μl of Cleaving master mix (from Step 1.11.1) to each sample. Gently mix by pipetting up and down 10 times.
1.11.5. Incubate the samples at 37°C for 15 minutes on a thermocycler with the thermocycler lid open.
1.11.6. Proceed immediately with the Post-reaction Wash (Section 1.11.7).
1.11.7. Post-reaction Wash
1.11.7.1. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.11.7.2. Add 150 μl of BW1 to each sample. Mix by pipetting up and down 10 times. Place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet.
1.11.7.3. Add 150 μl of BW2 to each sample. Mix by pipetting up and down 10 times.
![]() Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to Library Amplification in Section 1.12.
Samples can be kept at room temperature in BW2 buffer for up to 30 minutes. If longer storage is required, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Resuspend the beads in 100 μl of 1X TE. DNA-bound beads can be stored in 1X TE for up to 24 hours at 4°C. When ready to proceed with the protocol, place the samples on a magnet and wait for the solution to clear (~15 seconds). Remove the supernatant, and then remove the samples from the magnet. Add 150 μl of BW2 to each reaction, mix gently by pipetting up and down 10 times, and then proceed directly to Library Amplification in Section 1.12.
1.12. Library Amplification
Note: Refer to Chapter 2 if using the NEBNext Direct Index Primer Mix Plate provided in NEB #E7000X. Refer to Chapter 3 for guidelines on index pooling.
1.12.1. Place the reactions on a magnet, wait for the solution to clear (~15 seconds), remove the supernatant then remove the reactions from the magnet.
1.12.2. Add 45 μl of nuclease-free, molecular grade water to each reaction. Mix gently by pipetting up and down 10 times to completely resuspend the beads.
1.12.3. Add the following components to a sterile strip tube/well in a PCR plate:
| REAGENT | PER REACTION |
|---|---|
 (blue) Q5 Master Mix (blue) Q5 Master Mix |
50 μl |
 (blue) Index Primer Mix (blue) Index Primer Mix |
5 μl |
| Resuspended beads (from Step 1.12.2) | 45 μl |
| Total | 100 μl |
Gently mix by pipetting up and down 10 times. Seal the PCR plate or cap tubes.
1.12.5. Run the following program with the heated lid set to 105°C and place the samples in the thermocycler when the block temperature reaches 98°C:
| CYCLE STEP | TEMP | TIME | CYCLES |
|---|---|---|---|
| Initial Denaturation | 98°C | 30 seconds | 1 |
| Denaturation Annealing Extension | 98°C 62°C 72°C |
10 seconds 15 seconds 20 seconds |
20–25* |
| Final Extension | 72°C | 5 minutes | 1 |
| Hold | 4°C | ∞ |
*Follow the PCR cycle number recommendations listed in Table 1.12.1.
| INPUT DNA | RECOMMENDED NUMBER OF PCR CYCLES |
|---|---|
| 1,000 ng | 20 |
| 500 ng | 21 |
| 100 ng | 23 |
| 10 ng | 25 |
1.12.6. Proceed to Purify and Size Select Amplified Fragments in Section 1.13.
PCR reactions with beads can be stored for up to 24 hours at 4°C.
1.13. Purify and Size Select Amplified Fragments
1.13.1. If you detect significant evaporation from the PCR reaction, bring the volume up to 100 μl with molecular grade water.
1.13.2. Vortex the ![]() (brown) Sample Purification Beads to resuspend.
(brown) Sample Purification Beads to resuspend.
1.13.3. Add 85 μl of Sample Purification Beads to the PCR reactions. Mix well by pipetting up and down at least 10 times.
1.13.4. Incubate for 10 minutes uncapped at room temperature.
1.13.5. Place the tubes/PCR plate on a magnet. After the solution is clear (about 2 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain the DNA targets (Caution: do not discard beads).
1.13.6. Add 200 μl of freshly prepared (same day) 80% EtOH while the tubes/plate are on the magnet. Incubate at room temperature for 30 seconds and then carefully remove and discard the supernatant.
1.13.7. Repeat Step 1.13.6 once for a total of 2 washes in 80% EtOH, ensuring that all of the supernatant is removed from each reaction.
1.13.8. Incubate the samples, uncapped (or unsealed), at 37°C for 5 minutes on a thermocycler with the thermocycler lid open to dry the beads.
1.13.9. Remove the tubes/plate from the thermocycler and resuspend the dry beads in 102 μl of water. Incubate for 2 minutes at room temperature.
1.13.10. Place the tubes/plate on a magnet and allow the solution to clear (about 2 minutes).
1.13.11. Transfer 100 μl of the eluted library to fresh tubes/plate and add 85 μl of Sample Purification Beads. Mix well by pipetting up and down at least 10 times.
1.13.12. Incubate for 10 minutes at room temperature.
1.13.13. Place the tubes/plate on a magnet. After the solution is clear (about 2 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain the DNA targets (Caution: do not discard beads).
1.13.14. Add 200 μl of freshly prepared (same day) 80% EtOH while the tubes/plate are on the magnet. Incubate at room temperature for 30 seconds and then carefully remove and discard the supernatant.
1.13.15. Repeat Step 1.13.14 once for a total of 2 washes in 80% EtOH, ensuring that all of the supernatant is removed from each well.
1.13.16. Incubate the samples, uncapped (or unsealed), at 37°C for 2 minutes on a thermocycler with the thermocycler lid open to dry the beads.
1.13.17. Remove the tubes/plate from the thermocycler and resuspend the dry beads in 30 μl of 1X TE by gently pipetting (or gently vortex capped tubes/sealed plate, followed by a quick spin). Incubate for 2 minutes at room temperature.
1.13.18. Place the tubes on a magnet and allow the solution to clear (about 2 minutes).
1.13.19. Transfer 28 μl of the eluted library to a fresh tube.
1.13.20. Evaluate 1 μl of the eluted library on a High Sensitivity Bioanalyzer Chip or with a similar assay.