Getting started with loop-mediated isothermal amplification (LAMP)
Posted on Wednesday, October 12, 2022
By
Topic: Tips for the lab, What is Trending in Science
If you’ve wondered if loop-mediated isothermal amplification (LAMP) has the right sensitivity, specificity, rapidity, and robust high-throughput technical capabilities for your project, but you still have questions, we have answers. In this post, we’ve compiled some of the most frequently asked questions about the utility of LAMP, and throughout, we refer you to our valuable tools and resources to help you get started with LAMP.
(1) What is LAMP?
LAMP is a nucleic acid amplification technique with a simple workflow that is used to detect DNA or RNA inputs. LAMP takes advantage of the strand-displacing properties of Bst DNA polymerase (NEB #M0374) in conjunction with a unique primer design to allow rapid amplification of a specific sequence at one temperature. It is an ideal technique for field, point-of-care, or low-resource settings because sophisticated molecular diagnostic equipment is not required.
(2) How is LAMP different from PCR?
Both techniques amplify DNA, but there are fundamental differences that you should be aware of before embarking on a LAMP journey.
Here are some of the ways LAMP differs from PCR:
The reaction temperature – PCR requires cycling between temperatures to denature the DNA, anneal primers, and extend the new strand. LAMP uses one static temperature (65°C). LAMP’s strand displacing polymerase eliminates the need for the high (95°C) temperatures required for double-stranded DNA denaturation because it moves the top strand out of the way as it polymerizes.
The generated product - PCR creates many copies of discrete, single-sized amplicons. LAMP, on the other hand, creates long concatemers of the same repeated sequence. These form because LAMP uses six primers and a unique dumbbell structure that capitalizes on multiple priming points. The concatemers can be double or single-stranded, contain loops and branches, and can be hundreds of times longer than the amplified target sequence length.
The duration of the reaction - The timeframe of LAMP vs. PCR reactions is also quite different. Most PCR thermocycling protocols take upwards of 1.5 hours, particularly if incorporating real-time fluorescent measurements, while most LAMP reactions are complete in 30-40 minutes.
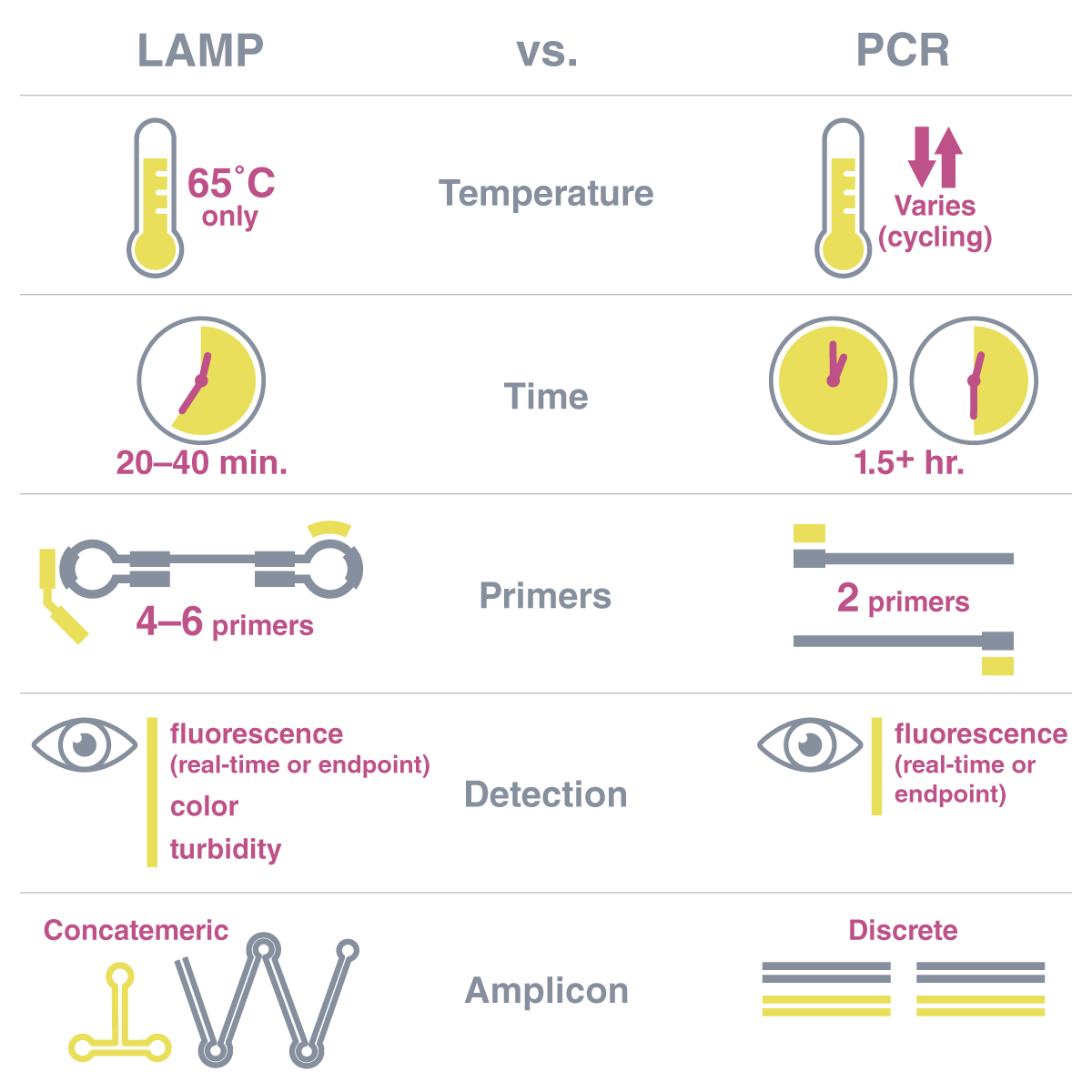
A comparison of some of the key differences between LAMP and PCR
(3) Why are there so many primers?
A primary goal of LAMP is to detect the target sequence as quickly as possible, and the multiple primers facilitate rapid amplification and expeditious detection.
LAMP uses four 'core' primers that are necessary for amplification to occur and two additional 'loop' primers that serve to accelerate the reaction. The core primers generate DNA that contains two regions of inverted self-complementarity. This forms the self-hybridizing loop structure at both ends of the target sequence leading to the formation of a “dumbbell” structure. The structure contains multiple opportunities for initiating synthesis, and the strand displacing Bst DNA Polymerase uses these priming points, resulting in rapid exponential amplification. This generates the concatemers mentioned above – long repeats of the short target sequence in a concise amount of time.
The video below gives a more detailed look at how a target sequence is primed for LAMP.

While the four core primers are necessary, the two loop primers are optional. Still, they are beneficial for speeding up the reaction and detecting the amplification signal sooner – the sooner the signal comes up, the greater the sensitivity of the reaction. The loop primers sit between two other sets of primers, so if there is not enough space, they can be challenging to design, though even one loop primer will help. It is recommended to try at least a couple of primer sets to see which gives the fastest amplification of the target with the least amplification in the no-template control (NTC) reactions.
Designing LAMP primers can seem daunting, but our new, straightforward LAMP Primer Design web tool makes this easy. We even have an instructional video tutorial to make designing your primers easier.
(4) What detection methods are used for LAMP reactions?
LAMP has various detection options, including fluorescence, endpoint, visual (colorimetric), and lateral flow. The most commonly used are fluorescence and visual detection.
Fluorescence Detection: LAMP can be monitored in real time or at the endpoint via fluorescence using an intercalating dye. NEB offers two LAMP kits that come supplied with this dye (NEB #E1700 and #E1708), and they can be monitored in a fluorimeter or qPCR machine for real-time detection, or a plate reader for endpoint detection. If monitoring in real time with intercalating dye, there is the option to include a melt curve which can help assess any nonspecific amplification. For applications requiring multiplex detection, various probe-based fluorescence methods can be used.
Visual Detection: LAMP can also be visually monitored with pH-based colorimetric output. In a weakly buffered solution using a pH-sensitive dye, this output is particularly useful for point-of-need testing. Detection by eye is, of course, very simple. Still, the strong color change of the phenol red dye can be measured quantitatively with a smartphone camera and image analysis or by absorbance measurement in a spectrophotometer or similar device. This can be useful for higher-throughput or diagnostic tests where user judgment is not required. Other non-pH-based colorimetric indicators like hydroxynaphthol blue or calcein can be used with our standard LAMP mixes if samples are likely to be strongly buffered or highly variable in pH.
(5) What sample types are compatible with LAMP? And how do I process the samples?
LAMP can accommodate a wide array of sample types and is generally more inhibitor tolerant than PCR. The best sensitivity is with the extracted nucleic acid; however, saliva samples, leaf punches, and samples in transport media can also be used.
Something to consider with colorimetric LAMP is that because it’s pH-based, it has more limitations with regard to sample type and quantity as the readout relies on the pH not being swung too far in one direction or the other, so more optimization on sample quantity is required. If the sample is in water, it is less of a problem, but if it’s in a tris buffer, for example, the amount of buffer in the reaction will need to be considered when establishing a reaction protocol. With samples of variable pH, like saliva, normalization through a lysis buffer or similar, can be helpful, and we do this ourselves for NEB’s in-house COVID testing program that uses colorimetric LAMP. Overall, LAMP is very flexible with regard to sample type. Conveniently, it can be used directly without undergoing a nucleic acid extraction method for field applications.
(6) What about high-throughput LAMP and carryover prevention?
As with any nucleic acid amplification technique, LAMP doesn’t need much material to get started. In a situation where there is only a minimal amount of starting material, the same target is repeatedly amplified, and there is an increased potential for an unintended amplification product from previous rounds of testing to contaminate the new reactions and serve as the substrate of a subsequent reaction. This can result in false positives. If reaction tubes are opened (for example, if the product will be run on a gel or used in a downstream application), it’s easy to get contamination, so incorporating some carryover contamination prevention is recommended, especially in a higher throughput setting.
Some of our LAMP kits (NEB #M1804/E1708/M1708) include dUTP and thermolabile UDG (Uracil DNA Glycosylase). Any contaminating dU-containing amplification products from previous reactions will be substrates for UDG activity, which will excise the uracil base, leaving poor substrates for LAMP amplification and reducing the possibility of carryover contamination.
(7) When is it better to use a LAMP assay instead of a qPCR assay?
LAMP is ideal if a simple ‘yes’ or ‘no’ answer is required, such as in a diagnostic assay. It gives a fast, qualitative answer. If you need any quantitation, qPCR is a more appropriate option. LAMP, particularly visual detection-based methods, is also more accessible in point-of-care and low-resource environments or if you want to test outside a lab with portable instruments.
(8) What are some of the applications LAMP has been used for?
LAMP is ideal in field applications, particularly where the transport of samples to a specialized lab is costly or geographically not feasible. Time-sensitive restrictions may also exist, whereby expeditious results are essential to productivity.
LAMP can be used in agriculture. Plant viral pathogens can devastate crops. The ability to test for a pathogen on-site, directly from a leaf punch, means that diagnosis is quick and less expensive than sending it to a specialized lab. Likewise, male cannabis plants can be identified early and removed from the desired crop. Again, a leaf punch is all that is required. There is minimal processing, and it’s a quick, cost-effective, high-throughput answer.
LAMP is also extremely useful in diagnosing and surveillance of neglected tropical diseases in low-resource settings. Additionally, field surveillance of vectors such as ticks and mosquitoes that harbor infectious diseases allows for immediate answers that help localize infected populations.
RT-LAMP has been used to diagnose SARS-CoV-2 in a point-of-care kit-based format and NEB®’s CLIA lab.
Basically, LAMP is useful in any study, diagnostic or surveillance situation where it would be beneficial not to rely on sophisticated equipment.
If you have more questions about LAMP methods and whether they would be appropriate for your project, reach out to NEB Technical Support.
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. See www.neb.com/trademarks. The use of these products may require you to obtain additional third-party intellectual property rights for certain applications. For more information, please email busdev@neb.com.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.


