
Harvesting improved methylome sequencing accuracy
Posted on Monday, December 14, 2020
By
Topic: What is Trending in Science
Plant epigenetics research is an attractive approach to enhance crops, rooted to accurate methylome analysis in a complex context. The performance of the Enzymatic Methyl-seq method merits replacing bisulfite conversion for high resolution plant methylation studies. Recently a team from the University of California Los Angeles took this leading-edge epigenomics method to task, comparing it to whole genome bisulfite sequencing (WGBS) results with Arabidopsis thaliana samples. Discover how researchers can improve studies of any eukaryotic methylome and you might become an EM-seq convert.
Appreciating crop developers
When we enjoy a good meal how often do we appreciate the origin of our food? As a bit of an archeology nerd, I find it fascinating to consider how ancient Native American societies used selective breeding over many centuries to produce remarkably nutritious crops like maize, potatoes, cassava, chocolate, beans, squash, tomatoes and peppers. The collective efforts of these ancient crop breeders produced around three-fifths of the crops now in cultivation - a fact seldom noted around dinner tables. In a corresponding manner, the work done by biologists today to enhance crops deserves appreciation. Plant genomics research has important implications to reduce hunger, protect the environment, and improve quality of life for everyone. It's a marvel how molecular biology can be applied to accelerate plant breeding.
The plant methylation approach
Many plant biologists consider methylation an appealing approach to improve agricultural plants that might have otherwise been developed through traditional plant breeding techniques. You can read more in this open access Argonomy review by Mercé et al. (2020) Induced Methylation in Plants as a Crop Improvement Tool: Progress and Perspectives. Some current research goals include crop drought tolerance, disease resistance and higher yields.
Scientists conducting methylome studies in the Arabidopsis thaliana plant model system find it reliable for developing molecular insights for crop enhancements and much more. Plant epigenetics pathways in general are more diverse than animal systems. Plant methylomes illustrate this fact by showing methylation in CG and non-CG contexts. These sites include CG, CHG and CHH contexts, (where H can be A, T, or C), and the asymmetric CHH sequence context (where H is A, T or C). Even given this increased complexity, studies using the Arabidopsis thaliana plant as a model system have reliably generated critical knowledge for both plant and animal genetics. Around 88% of Arabidopsis papers get referenced by non-Arabidopsis papers. Apparently it has been quite useful that this organism can tolerate mutations that nearly eliminate methylation. Plasticity is a virtue for science. Methylome studies using Arabidopsis have anticipated exciting tools to bring the best natural traits out of crops, as well as extending our understanding of conserved epigenetic phenomena that are relevant to human health.
The question is can we improve the accuracy and reliability of plant methylome studies in general?
Based on a new report by UCLA researchers comparing the Enzymatic Methyl-seq method against whole genome bisulfite sequencing, the answer is yes.
The cons of bisulfite conversion
First off, please forgive me because I am going to begin by explaining the drawbacks of a substance that has provided tremendous epigenomics insights. Observed in its isolated state, sodium bisulfite / NaHSo3 is a chemical solid with acute toxicity to skin, eyes and lungs, with the “bonus” property of wafting the scent of rotten eggs.
Sound attractive? Not really.
However, it IS useful. Applications include treating drinking water, preventing corrosion, wine making, as a food preservative, and my personal favorite; its use in powerful epigenomics methods, as described in the seminal paper by Frommer et al., 1992. Bisulfite-seq has been the foundational method for methylome studies, bar none. Back in 2008, Hikoya Hayatsu, one of the two discoverers of this chemical, offered a personal account of its use in DNA methylation analysis, referring to bisulfite as a “fascinating” agent with “mysterious” actions. (I love that sort of rare prose found in scientific reports.) It’s fair to add the descriptive term “influential”, since the impact of bisulfite-seq has been tremendous in so many areas of epigenetics research.
The value of NaHSO3 to genomics is its property of selectively converting unmethylated cytosine in DNA into uracil, while leaving methylated and hydroxy methylated CpG dinucleotides unchanged. This is followed up with pcr using a DNA Polymerase that will amplify the uracil as thymine. Q5U® Hot Start High-fidelity DNA Polymerase is a very good choice (NEB #M0515), because no initial activation time is required, and the buffer composition is optimized for AT-rich pcr. Simply put, the transformation looks like this: ATCG → ATUG → ATTG. Bisulfite treated libraries can then be compared to a reference genome to show methylation status at C/T positions with single base pair resolution. Finally, you get to go figure out what all your data means in your study scenario.
Now come the cons. WGBS protocols can suffer from multiple biases and variable performance. The bisulfite conversion protocol itself is toxic and cumbersome. It requires high temperature and acidic or alkaline pH in different protocol stages. Upfront 90% of the input DNA can be degraded from bisulfite conversion. From there the negative impacts build. Since CG regions take greater damage than AT regions, fragments from these regions are less successfully incorporated into libraries, and thus less amplified. This results in AT rich libraries that are not reflective of your original samples.
Unsurprisingly, DNA damage is a salient factor in experimental results. The paper Nelly Olova et al. (2018) Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data from Wolf Reik group reports that up to 20% of DNA methylation changes of their reviewed WGBS datasets were not actually biologically based. Ug! It sounds like the drawbacks of bisulfite conversion methods could be factoring into the scientific reproducibility crisis.
Enzymatic Methyl-seq
In comparison to bisulfite-seq, Enzymatic Methyl-seq or “EM-seq” is a cost-effective greener chemistry method that produces more accurate and informative methylome data. It's a high-resolution technique for discriminating cytosine and methylated cytosine residues, without losing 90% of input DNA. You get longer inserts, lower duplication rates, and a higher percentage of mapped reads, with less GC bias. Using enzymes to convert DNA in place of bisulfite treatment recovers more amplifiable DNA as input for NGS, with minimal GC bias. You can use the same bioinformatics tools because methylated cytosine appears as cytosine and unmethylated cytosine appears as thymine in the end. This is a user-friendly method designed to improve methylome data.
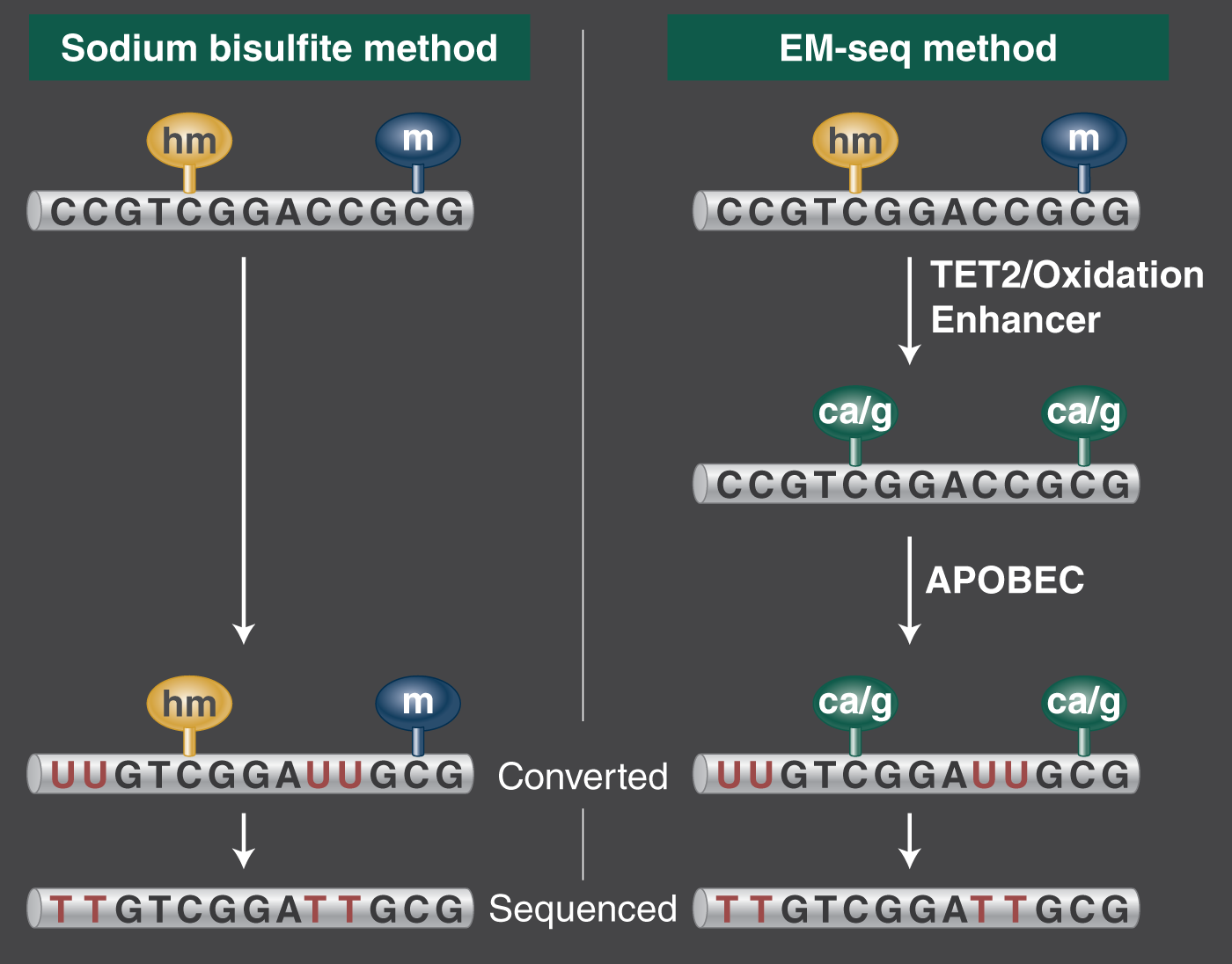
EM-seq is more reliable and accurate than WGBS
Scientists focused on the perceived reliability of using bisulfite conversion as the gold standard, can consider new evidence that EM-seq is more reproducible than WGBS. Principal investigator Steven Jacobsen and his team from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA took this leading-edge methylation analysis method to task with in the paper Epigenetics & Chromatin, Suhua Feng, Zhenhui Zhong, Ming Wang & Steven E. Jacobsen (2020) Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis thaliana with enzymatic methyl sequencing. The authors report on the advantages with EM-seq over whole genome bisulfite sequencing (WGBS) based on a detailed analysis of DNA damage, false positives due to non-conversion, uneven and missing coverage, and biased representation of methylated versus unmethylated DNA in final libraries. EM-seq was more consistent within libraries in all examined quality aspects and in overall methylation detection.
- EM-seq has longer inserts, a higher mapping rate, lower duplication rate, and lower false-positive rate than WGBS.
- Coverage is less affected by GC content than WGBS.
- Reproducibility makes EM-seq more desirable than WGBS, especially for big data projects that require integration of datasets obtained across a wide variety of source materials, locations and time points, and processed by personnel with different levels of expertise.
- Low background coupled with high reproducibility makes EM-seq suitable for projects aimed at revealing subtle methylation changes in different sample types and conditions.
- The results format is convenient for bioinformatics
It’s wonderful to see feedback that speaks to the accuracy and reliability of a method developed here by New England Biolabs scientists! There are so many other eukaryote sample types that will benefit from more accurate genome methylation detection. Stay tuned for updates!
NEB will not rent, sell or otherwise transfer your data to a third party for monetary consideration. See our Privacy Policy for details. View our Community Guidelines.
Don’t miss out on our latest NEBinspired blog releases!
- Sign up to receive our e-newsletter
- Download your favorite feed reader and subscribe to our RSS feed
Be a part of NEBinspired! Submit your idea to have it featured in our blog.



