Protocol for use with NEBNext Ultra DNA Library Prep Kit for Illumina (E7370)
Symbols
| This caution sign signifies a step in the protocol that has multiple paths leading to the same end point but is dependent on a user variable, like the amount of input DNA. |
|
| Colored bullets indicate the cap color of the reagent to be added to a reaction. |
Starting Material: 5 ng–1 µg fragmented FFPE DNA.
2.1. NEBNext FFPE Repair
2.1.1. Mix the following components in a sterile nuclease-free tube:
| FFPE DNA | 53.5 µl |
 (green) FFPE DNA Repair Buffer (green) FFPE DNA Repair Buffer |
6.5 µl |
 (green) NEBNext FFPE DNA repair Mix (green) NEBNext FFPE DNA repair Mix |
2 µl |
| Total Volume | 62 µl |
2.1.2. Mix by pipetting followed by a quick spin to collect all liquid from the sides of the tube.
2.1.3. Incubate at 20°C for 15 minutes.
2.2. Cleanup Using AMPure XP Beads
2.2.1.Vortex AMPure XP Beads to resuspend.
2.2.2. Add 186 μl (3X) of resuspended AMPure XP Beads to the repair reaction. Mix thoroughly on a vortex mixer or by pipetting up and down at least 10 times.
2.2.3. Incubate for 5 minutes at room temperature.
2.2.4. Put the tube/PCR plate on an appropriate magnetic stand to separate beads from supernatant. After the solution is clear (about 5 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain the DNA targets.
2.2.5. Add 200 μl of 80% freshly prepared ethanol to the tube/PCR plate while in the magnetic stand. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
2.2.6. Repeat Step 2.2.5 once.
2.2.7. Air dry beads for up to 5 minutes while the tube/PCR plate is on the magnetic stand with the lid open.
Caution: Do not over-dry the beads. This may result in lower recovery of DNA target. Elute the samples when the beads are still dark brown and glossy looking, but when all visible liquid has evaporated. When the beads turn lighter brown and start to crack, they are too dry.
2.2.8. Remove the tube/plate from the magnet. Elute DNA target by adding 60 μl 0.1X TE to the beads. Mix well on a vortex mixer or by pipetting up and down, and incubate for 2 minutes at room temperature. Put the tube/PCR plate in the magnetic stand until the solution is clear.
2.2.9. Without disturbing the bead pellet, carefully transfer 55.5 μl of the supernatant to a fresh, sterile microfuge tube.
2.3. NEBNext End Prep
2.3.1. Add the following directly to the FFPE repair reaction mixture:
 (green) End Repair Reaction Buffer (10X) (green) End Repair Reaction Buffer (10X) |
6.5 µl |
 (green) End Prep Enzyme Mix (green) End Prep Enzyme Mix |
3.0 µl |
| Repaired DNA from above (Step 2.2.9) | 55.5 µl |
| Total Volume | 65 µl |
2.3.2. Mix by pipetting at least 10 times followed by a quick spin to collect all liquid from the sides of the tube.
2.3.3. Place in a thermocycler, with the heated lit set to 75°C, and run the following program:
30 minutes @ 20°C
30 minutes @ 65°C
Hold at 4°C
2.4. Adaptor Ligation
![]() If DNA input it < 100 ng, dilute the NEBNext Adaptor for Illumina (provided at 15 µM) 10 fold in 10 mM Tris-HCl pH 7.5 with 10 mM NaCl to a final concentration of 1.5 µM; use immediately.
If DNA input it < 100 ng, dilute the NEBNext Adaptor for Illumina (provided at 15 µM) 10 fold in 10 mM Tris-HCl pH 7.5 with 10 mM NaCl to a final concentration of 1.5 µM; use immediately.
2.4.1. Add the following components directly to the End Prep readction mixture:
 (red) Blunt/TA Ligase Master Mix (red) Blunt/TA Ligase Master Mix |
15 µl |
 (red) NEBNext Adaptor for Illumina (15 µM)* (red) NEBNext Adaptor for Illumina (15 µM)* |
|
 (red) Ligation Enhancer (red) Ligation Enhancer |
1 µl |
| End Prep DNA mixture from above (Step 2.3.3) | 65 µl |
| Total Volume | 83.5 µl |
*The NEBNext adaptor is provided in NEBNext Singleplex or Multiplex Oligos for Illumina.
2.4.2. Mix by pipetting followed by a quick spin to collect all liquid from the sides of the tube.
2.4.3. Incubate at 20°C for 15 minutes in a thermocycler.
2.4.4. Add 3 µl of  (red) USER Enzyme to the ligation mixture from Step 2.4.3. Mix well and incubate at 37°C for 15 minutes.
(red) USER Enzyme to the ligation mixture from Step 2.4.3. Mix well and incubate at 37°C for 15 minutes.
Note: This step is for use with NEBNext adaptors only. USER Enzyme can be found in the NEBNext Singleplex or Multiplex Oligos for Illumina.
![]() A precipitate can form upon thawing of the NEBNext Q5 Hot Start HiFi PCR Master Mix. To ensure optimal performance, place the master mix at room temperature while performing cleanup of adaptor–ligated DNA. Once thawed, gently mix by inverting the tube several times.
A precipitate can form upon thawing of the NEBNext Q5 Hot Start HiFi PCR Master Mix. To ensure optimal performance, place the master mix at room temperature while performing cleanup of adaptor–ligated DNA. Once thawed, gently mix by inverting the tube several times.
2.5. Cleanup of Adaptor–Ligated DNA
2.5.1. Vortex AMPure XP Beads to resuspend.
2.5.2. Add 86.5 µl resuspended AMPure XP Beads to the ligation reaction. Mix well by pipetting up and down at least 10 times.
2.5.3. Incubate for 5 minutes at room temperature.
2.5.4. Quickly spin the tube and place it on an appropriate magnetic stand to separate beads from supernatant. After the solution is clear (about 5 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets (Caution: do not discard the beads).
2.5.5. Add 200 µl of 80% freshly prepared ethanol to the tube while in the magnetic stand. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
2.5.6. Repeat Step 2.5.5 once.
2.5.7. Air dry the beads for up to 5 minutes while the tube is on the magnetic stand with the lid open.
Caution: Do not over-dry the beads. This may result in lower recovery of the DNA target. Elute the samples when the beads are still dark brown and glossy looking, but when all visible has evaporated. When the beads turn lighter brown and start to crack, they are too dry.
2.5.8. Remove the tube/plate from the magnet. Elute the DNA target from the beads by adding 22 µl of 10 mM Tris-HCl, pH 8.0 or 0.1X TE.
2.5.9. Mix well by pipetting up and down at least 10 times, or on a vortex mixer.
2.5.10. Quickly spin the tube or incubate for 2 minutes at room temperature.
2.5.11. Place the tube on a magnetic stand. After the solution is clear (about 5 minutes), transfer 20 µl to a new PCR tube for amplification.
2.5.12. Proceed to PCR Amplification.
2.6. PCR Amplification
![]() Check and verify that the concentration of your oligos is 10 µM on the label.
Check and verify that the concentration of your oligos is 10 µM on the label.
![]() Use Option A for any NEBNext oligos kit where index primers are supplied in tubes. These kits have the forward and reverse primers in separate tubes.
Use Option A for any NEBNext oligos kit where index primers are supplied in tubes. These kits have the forward and reverse primers in separate tubes.
Use Option B for any NEBNext oligos kit where index primers are supplied in a 96-well plate format. These kits have the forward and reverse (i7 and i5) primers combined.
2.6.1A. Forward and Reverse Primer Not Already Combined
Mix the following components in sterile strip tubes:
| Adaptor Ligated DNA Fragments (from Step 2.5.11) | 15 µl |
  (blue) Index Primer/i7 Primer*,** (blue) Index Primer/i7 Primer*,** |
5 µl |
  (blue) Universal PCR Primer/i5 Primer*,** (blue) Universal PCR Primer/i5 Primer*,** |
5 µl |
  (blue) NEBNext Q5 Hot Start HiFi PCR Master Mix (blue) NEBNext Q5 Hot Start HiFi PCR Master Mix |
25 µl |
| Total Volume | 50 µl |
*NEBNext Oligos must be purchased separately from the library prep kit. Refer to the corresponding NEBNext Oligo kit manual for determining valid barcode combinations.
**Use only one i7/index primer per sample. Use only one i5 primer (or the universal primer for single index kits) per sample.
2.6.1B. Forward and Reverse Primer Already Combined
Mix the following components in sterile strip tubes:
| Adaptor Ligated DNA Fragments (from Step 2.5.11) | 15 µl |
  (blue) Index (X)/Universal Primer Mix* (blue) Index (X)/Universal Primer Mix* |
10 µl |
  (blue) NEBNext Ultra II Q5 Master Mix (blue) NEBNext Ultra II Q5 Master Mix |
25 µl |
| Total Volume | 50 µl |
*NEBNext Oligos must be purchased separately from the library prep kit. Refer to the corresponding NEBNext Oligo kit manual for determining valid barcode combinations.
2.6.2. PCR cycling conditions:
| Cycle Step | Temp | Time | Cycles |
| Initial Denaturation | 98°C | 30 seconds | 1 |
| Denaturation | 98°C |
10 seconds | 4-15* |
| Annealing/Extension | 65°C | 75 seconds | |
| Final Extension | 65°C | 5 minutes | 1 |
| Hold | 4°C | ∞ |
*We suggest 4–6 cycles for 1 µg DNA input, 8–9 cycles for 100 ng, 11–12 cycles for 20 ng, and 13–15 cycles for 5 ng. Further optimization of PCR cycle number may be required.
2.6.3. Proceed to cleanup of PCR Amplification
2.7. Cleanup of PCR Amplification
2.7.1. Vortex AMPure XP Beads to resuspend.
2.7.2. Add 50 µl of resuspended AMPure XP Beads to the PCR reactions (~50 µl). Mix well by pipetting and and down at least 10 times.
2.7.3. Incubate for 5 minutes at room temperature.
2.7.4. Quickly spin the tube and place it on an appropriate magnetic stand to separate beads from the supernatant. After the solution is clear (about 5 minutes), carefully remove and discard the supernatant. Be careful not to disturb the beads that contain DNA targets (Caution: Do not discard the beads).
2.7.5. Add 200 µl of 80% ethanol to the PCR plate while in the magnetic stand. Incubate at room temperature for 30 seconds, and then carefully remove and discard the supernatant.
2.7.6. Repeat Step 2.7.5 once.
2.7.7. Air dry the beads for up to 5 minutes while the PCR plate is on the magnetic stand with the lid open.
Caution: Do not over-dry the beads. This may result in lower recovery of DNA target. Elute the samples when the beads are still dark brown and glossy looking, but when all visible liquid has evaporated. When the beads turn lighter brown and start to crack, they are too dry.
2.7.8. Remove the tube/plate from the magnet. Elute DNA target from the beads into 33 µl of 0.1X TE. Mix well by pipetting up and down at least 10 times. Quickly spin the tube and place it on an appropriate magnetic stand to separate beads from supernatant. After the solution is clear (about 5 minutes), carefully transfer 28 µl supernatant to a new PCR tube. Libraries can be stored at –20°C.
2.7.9. Dilute 2–3 µl of the library 5-fold with 10 mM Tris or 0.1X TE and check the size distribution on Agilent Bioanalyzer® (High Sensitivity chip)
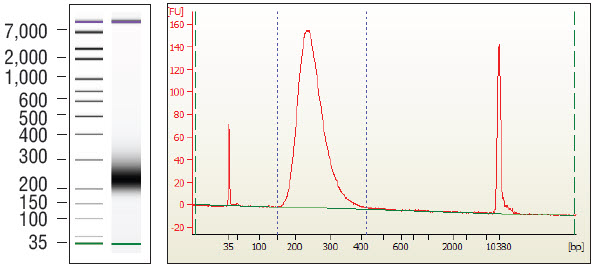
2.7.10. A sharp peak at 128 bp corresponds to adaptor-dimer. We recommend repeating 2.7.1 to 2.7.9 if this occurs.
Figure 2.1: Example of a library prepared with normal human liver FFPE DNA.