The Next Generation of Cell-Free Protein Synthesis
Background of Cell-Free Protein Synthesis
Since the early pioneering work of Nirenberg and Matthaei in 1961 (1), which demonstrated in vitro protein translation using cell extracts, cell-free protein synthesis has become an important tool for molecular biologists by playing a central role in a wide variety of applications (2). In the post-genomic era, cell-free protein synthesis has the potential to become one of the most important high throughput technologies for functional genomics and proteomics.
The biggest advantage, compared to protein production in living cells, is that cell-free protein synthesis is the quickest way to obtain an expressed phenotype (protein) from a genotype (gene). Starting with a PCR or plasmid template, in vitro protein synthesis and functional assays can be carried out in a few hours. Moreover, it is independent of host cells. Proteins which are toxic or prone to proteolytic degradation can be readily prepared in vitro.
Commercially available cell-free protein synthesis systems are typically derived from cell extracts of Escherichia coli S30, rabbit reticulocytes or wheat germ. The drawback of extract-based systems is that they often contain nonspecific nucleases and proteases that adversely affect protein synthesis. In addition, the cell extract is like a “black box” in which numerous uncharacterized activities may modify or interfere with subsequent downstream assays.
Some of these limitations can be partially overcome, for instance, by using engineered strains or by adding various inhibitors. Nevertheless, the problems cannot be solved at the root level.
The "PURE" Approach
The first complete in vitro reconstitution of protein translation from E. coli was accomplished in 2001 by Dr. Takuya Ueda’s lab at the University of Tokyo. This became known as the “PURE” system, which stands for Protein Synthesis Using Recombinant Elements”(3). This system was then commercialized as the PURESYSTEM® by the Post Genome Institute (PGI) (Tokyo, Japan). Except for the ribosomes and tRNAs, which are highly purified from E. coli, the PURE system reconstitutes the E. coli translation machinery with fully recombinant proteins.
PURExpress™ from NEB is based on the PURESYSTEM® from PGI, and improves on the original “Classic II” Kit by optimizing components to increase yield of protein synthesis. For more information please click here.
The PURE system includes:- His-Tagged Translation Factors -Initiation Factors (IF1, IF2, IF3)
- E. coli Ribosomes
- E. coli tRNAs
- Energy Regeneration System
- NTPs, Amino Acids, Salts, Buffer
-Elongation Factors (EF-Tu, EF-Ts, EF-G)
-Release Factors (RF1, RF2, RF3)
-Ribosome Recycling Factor
-20 Aminoacyl tRNA synthetases
-Methionyl tRNA formyltransferase
In addition, recombinant T7 RNA polymerase is used to couple transcription to translation. The PURE system represents an important step towards a totally defined in vitro transcription/translation system, thus avoiding the “black box” nature of the cell extract-based systems.
Advantages and Applications of the PURE system
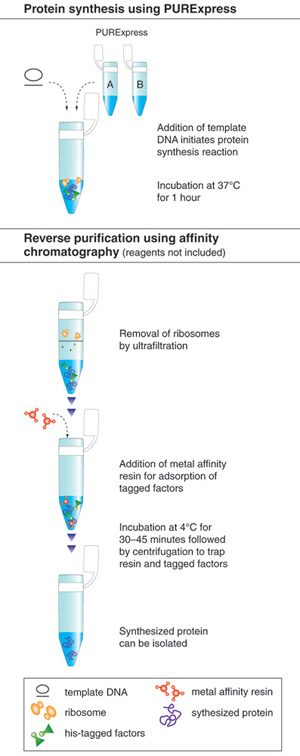
The PURE system is more robust and convenient than most extract-based systems for many in vitro applications. The immediate advantage is the significantly reduced level of all contaminating activities. It can be used to express a wide range of protein targets and has the capacity for a yield of more than 100 μg/ml. The activity of the synthesized protein can often be directly assayed without purification due to the low background activity of the translation mixture. All recombinant protein factors inside the PURE system are His-tagged, in some cases allowing the synthesized protein to be “reverse-purified” (Figure 1,2)(3).
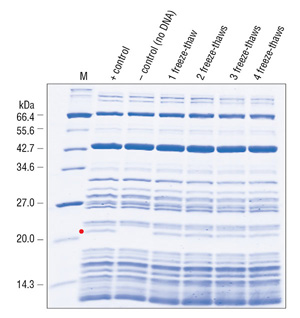
The purity of this system allows it to withstand more than five freeze-thaw cycles without losing its efficiency, further extending its shelf life (Figure 3)(Cantor, E., unpublished observation).
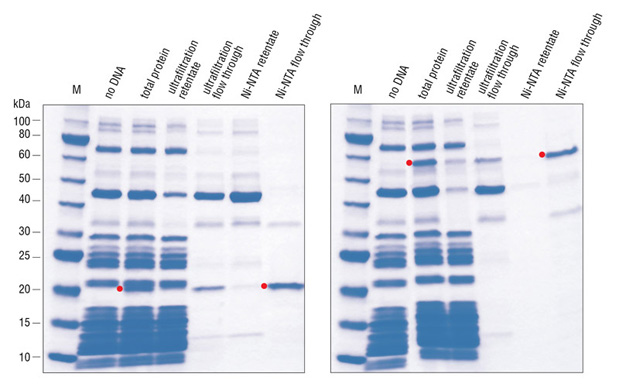
The advantages and applications of the PURE system have been demonstrated in various in vitro applications including:
1) High throughput functional genomics and proteomics
The simple format of the PURE system allows it to be easily integrated into high throughput platforms for functional genomics and proteomics studies. The absence of any nuclease activities ensures the stability of linear DNA templates during protein synthesis. Individual DNA templates for in vitro expression can be generated by PCR, eliminating the time-consuming cloning process. This feature is particularly useful for high throughput screening at the whole genome scale, either for novel activities or for protein-protein interactions. For structural genomics projects, the PURE system can be an alternative route to acquire difficult protein targets which resist traditional cellular expression (4).
2) Protein engineering
Directed evolution of proteins in vitro is a powerful tool for improving and creating biocatalysts. A number of in vitro evolution methodologies, such as mRNA display (5), ribosome display (6) and in vitro compartmentalization (7), depend on in vitro translation. For example, NEB first demonstrated that the PURE system is uniquely suited for the in vitro selection of restriction endonucleases using the in vitro compartmentalization method (8), as it is free of nonspecific nuclease activity. In this study, the PURE system and the DNA library were dispersed into more than 109 aqueous droplets in a water-in-oil emulsion. The droplet encapsulation provides a linkage between the phenotype (expressed protein) and the genotype (DNA), which sets the stage for the specific selection of restriction enzyme genes. Other researchers have reported that using the PURE system greatly improves the efficiency of ribosome display (9).
Systematically mutagenized protein-coding libraries can be used as well to test if a specific mutation(s) affects protein function. Again, only a few PCR steps are needed to obtain the mutant protein, providing a quick experimental verification of hypotheses.
3) Study of protein expression, translation and folding
The PURE system contains the minimal set of factors necessary for in vitro protein translation. It is largely free of chaperones and other cellular factors for post-translational modifications, thus providing a starting point to study the involvement of these factors in transcription/translation regulation and nascent chain folding (10).
It can also be used to produce “clean” proteins which, if purified from traditional cellular hosts, may come with undesired modifications or bound co-factors. A number of research labs studying translation routinely use home-made reconstituted systems to study different aspects of translation.
Read: Bypassing Common Obstacles in Protein Expression
4) Incorporation of unnatural amino acids
Another important advantage of the PURE system is the ability to control its composition. For example, omission of the release factor 1 (RF1) in the PURE system allows unnatural amino acids to be efficiently incorporated at specific amber codon sites via chemically mis-acylated suppressor tRNA (3,11). It was recently reported that the translation apparatus of E. coli can tolerate a wide range of amino acid derivatives, revealing even greater potential for the ribosomal synthesis of unnatural peptides using reconstituted systems (12).
The future of cell-free protein synthesis
Future generations of cell-free protein synthesis systems should contain defined components, providing a clean background and at the same time be able to produce correctly folded proteins of any type in high yields. The PURE system clearly represents a breakthrough towards this goal. Improved reconstituted systems will be customized for expression of complex proteins such as large membrane proteins, multi-subunit assemblies and specifically modified proteins etc.
Customization will also permit diversity in the protein synthesis conditions, such as elevated or decreased temperature, ionic strength and redox environment etc.
Protein translation is one of the core processes in living organisms that serve as a “central node” to network other biological processes. There is no doubt that creative applications of the reconstituted systems will go beyond producing proteins and into such diverse fields as synthetic biology, systems biology and medical diagnostics.
Reference
- Nirenberg, M.W. and Matthaei, J.H. (1961) The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA, 47, 1588–1602.
- Katzen, F., Chang, G. and Kudlicki, W. (2005) The past, present and future of cell-free protein synthesis. Trends. Biotechnol. 23, 150–156.
- Shimizu, Y., et al. (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755.
- Graslund, S., et al. (2008) Protein production and purification. Nat. Methods, 5, 135–146.
- Roberts, R.W. and Szostak, J.W. (1997) RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA, 94, 12297–12302.
- Hanes, J. and Pluckthun, A. (1997) in vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA, 94, 4937–4942.
- Tawfik, D.S. and Griffiths, A.D. (1998) Man-made cell-like compartments for molecular evolution. Nat Biotechnol. 16, 652–656.
- Zheng, Y. and Roberts, R.J. (2007) Selection of restriction endonucleases using artificial cells. Nucleic Acids Res. 35, e83.
- Villemagne, D., Jackson, R. and Douthwaite, J.A. (2006) Highly efficient ribosome display selection by use of purified components for in vitro translation. J. Immunol. Methods, 313, 140–148.
- Kaiser, C.M., et al. (2006) Real-time observation of trigger factor function on translating ribosomes. Nature, 444, 455–460.
- Noren, C.J., et al. (1989) A general method for site-specific incorporation of unnatural amino acids into proteins. Science, 244, 182–188.
- Hartman, M.C., et al. (2007) An expanded set of amino Acid analogs for the ribosomal translation of unnatural peptides. PLoS ONE, 2, e972.

