NEB® TV Ep. 16 – Overview and Applications of qPCR
Script
Deana Martin: NEB TV. What's trending in science.
Deana Martin: Welcome to NEB TV. Today I'm joined by Yan Xu who is an Application and Product Development Scientist here at NEB, and we are talking about qPCR.
Deana Martin: In our "Science in 60," Yan is going to give us an overview of qPCR, then we'll hear about a novel method that was developed by NEB researchers to analyze large amounts of qPCR data. This method was used in the development of our Luna® qPCR product portfolio. And lastly, we'll answer some questions we've received from customers on our Luna products.
Deana Martin: Are you ready?
Yan Xu: Yes.
Deana Martin: Great. Let's get started.
Yan Xu: In contrast to PCR where the detection of our target sequence occurs at the end of a reaction, qPCR monitors DNA amplification as the reaction progresses. In qPCR assays, reaction progression is monitored by an increase in fluorescence signal. The fluorophore can be a double-stranded DNA binding dye or covalently attached to our target-specific probe.
Yan Xu: In dye based assays, the dye fluoresces when bound to double-stranded DNA but displays much weaker fluorescence in the presence of single-stranded DNA. This results an increased fluorescence signal as the reaction progresses and double-stranded DNA is formed. In probe-based assays, fluorescence is generated by the separation of a dye quencher pair during amplification by cleavage of the probe or by other means of separation. Each method relies on the detection of a fluorescent signal or a background and determination of the cycle, Cq, where this occurred, allowing relative or absolute quantification of the target.
Janine Borgaro: We examined the Luna mixes, along with a variety of commercially available mixes on a broad set of targets following manufacturer recommendations.
Janine Borgaro: By including a five-log dilution of template and a no template control, all run in triplicate, 18 different qPCR curves regenerated per target. Each target was also run twice by two different users. Because this data set became quite large, we needed to create a better more scalable way to visualize our results.
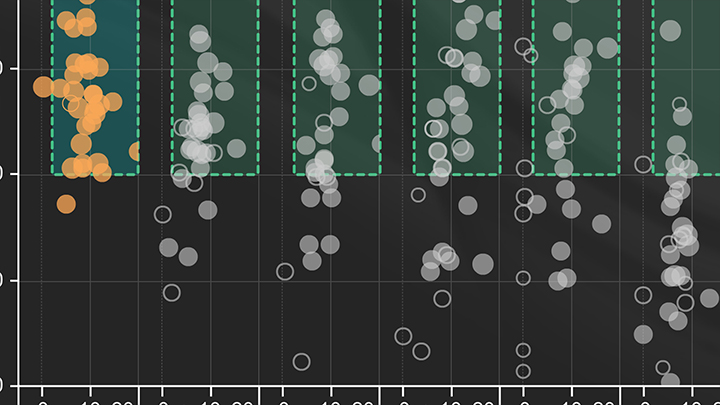
Janine Borgaro: We started by plotting the calculated efficiency of the standard curve against the Delta Cq which is the distance between the last template dilution and the no template control, thereby translating the 18 wells worth of data per target into a single dot. By providing guidelines around the typically accepted values for these parameters, for example, a 90-110% efficiency and a Delta Cq of three or greater, this created a graphical visual we refer to as dots and boxes. However, it didn't quite capture all the relevant details of each run. In order to represent additional information, the overall quality of the qPCR curves was scored on a scale from one to five. This scoring method was built upon previously published work by Hall et al, which encompassed factors such as curve sigmoidality and triplicate Cq tightness. The quality score was then used to define the size of the dot where the larger dot represented a higher quality score.
Janine Borgaro: This analysis method allowed us to quickly compare results across multiple targets or conditions and across a large data set in order to better observe trends. For example, here we see the data set that includes the Luna Universal qPCR Master Mix compared to a variety of other commercial mixes designed for DNA dye-based qPCR. For example, Cyber.
Janine Borgaro: On a total of 18 unique genomic and cDNA targets run in triplicate by two different users. Across these 36 experiments, we see the strong performance of the Luna mix for 86% of the experiments yielded high quality results that fall in the box. Similar results were observed for all Luna mixes and these dots and box visuals can be found on each of the individual product pages.
Janine Borgaro: We've run thousands of qPCR experiments and looked at curve after curve to make the best possible qPCR reagents. We hope the work that we did will enable the research you'll do.
Lynne Apone: One of the most common questions we receive about our Luna qPCR kits is whether or not they work with different qPCR machines or instruments. These kits have been designed specifically to work with a wide range of instruments. They can work in instruments that do not require a passive reference dye, those that require low concentration, passive reference dyes, or those that require high concentration, passive reference dyes. They can also be used with standard and fast cycling conditions.
Lynne Apone: Another common question we receive is "why is the Luna qPCR Mix blue?" So this mix contains an inert reference dye that gives it a clear blue color. This colored appearance makes it easier to identify which wells of a qPCR plate have been loaded with reagents. And this can be very difficult with the small well size and opaque nature of many 96- and 384- well qPCR plates. The blue dye has been studied extensively in both qPCR and PCR and it does not inhibit or interfere with qPCR detection.
Lynne Apone: Another common question we receive is whether the Universal Probe Master Mix can be used in multiplex reactions. The answer is yes. We have tested three amplicons successfully using the mix. You want to start by choosing different fluorophores that are detected in different channels on the qPCR instrument. Then you want to test each amplicon separately and individually in a singleplex reaction. When you multiplex them, if any one amplicon looks like it's deteriorating compared to singleplex results, then you want to optimize. One of the easiest things to optimize is primer concentration. So you can start by increasing the primer concentration for targets that are of low abundance and decreasing prier concentration for targets that have high abundance.
Lynne Apone: For additional tips on how to multiplex, please contact us at info@neb.com.
Deana Martin: So Yan, thanks so much for joining me today.
Yan Xu: Sure. Thank you.
Deana Martin: And thanks for watching. If you have any suggestions for future episodes, please let us know.