Standard Protocol for Restriction Enzyme Digests

Script
We're going to talk about setting up a standard restriction enzyme digestion today. Before you begin the setup of the actual reaction, it's a good idea to have an ice bucket available and all of the components to the reaction thawed to completion and on ice. One of the things that commonly gets missed is having the buffer completely thawed and mixed well before adding it to the reaction.
A standard reaction is 50 microliters. It calls for one microgram of your target DNA, five microliters of the restriction buffer, five to 10 units of enzyme, and then supplementing the rest of the 50 microliters with distilled water.
If your enzyme is a Time-Saver qualified enzyme, it will only require a 5 to 15 minute incubation period. If it's not, it will require an hour. It's a good idea once the reaction is complete that it's mixed well. Mixing can be done by cycling through a pipetman several times or by flicking it with your fingers. We recommend not vortexing digestion mix. If you do choose to flick with your fingers, I would recommend spinning the reaction briefly in the centrifuge before putting the reaction at its temperature.
Most digests of a microgram of DNA will require five to 10 units of enzyme. Genomic DNA, specifically mammalian DNA, may require longer incubations and may require more enzyme.
At the completion of the incubation period, you want to add loading dye and then load it directly onto your gel.
Related Videos
-

NEB® Restriction Enzyme Double Digest Protocol -
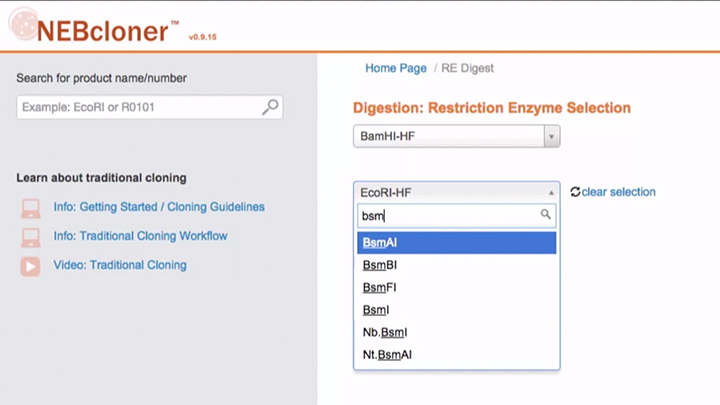
Double Digestion with NEBcloner -
How do restriction enzymes interact with DNA?

